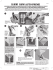XPrimer pression XPrimer pression
Transcription
XPrimer pression XPrimer pression
PREMIER Biosoft X pression Primer I n t e r n a t i o n a l Ap r att R1 Gene Neg sel att B1 A revolutionary tagged primer design tool for designing expression cloning experiments Km r att R2 att B2 Design thousands of primer pairs optimized for Gateway®, BD In-Fusion™, TOPO® Tools and epitope systems Add functionally useful tags for any expression system of your choice and design tagged primers Automatically maintain the reading frame of the ORF to be amplified when adding tags View primer secondary structures graphically Automatic check for in-frame termination Design sequencing primers for multiple sequences in a single search run Sequencing primers are designed across amplicons with a user defined interval between forward primers. Reverse primers stagger on the opposing strands Design primers for in vitro transcriptiontranslation Primer Tm is calculated using highly accurate SantaLucia nearest neighbor thermodynamic values Optimizes all primers in a single search run for uniform PCR cycling conditions Primers are screened for thermodynamic properties and secondary structures Retrieve batches of ORF sequences from Entrez using accession or GI numbers Comprehensive project management for easy and convenient storage and access of data from multiple experiments Extensive Assay Support for Tagged Primer design Use the sophisticated algorithm of Xpression Primer to design thousands of tagged primers for expression cloning systems such as Gateway®, BD In-Fusion™, epitope and TOPO® Tools. You can choose to amplify an entire ORF or generate N terminal or C terminal fusion proteins. Xpression Primer ensures that the reading frame of the amplified ORF is conserved. To work with other expression systems, simply add functional tags of your choice and design tagged primers. Successful Amplification with Nested PCR To ensure the success of your PCR experiment, let Xpression Primer design nested tagged primers to amplify ORFs. You can locate the outer primers anywhere in the UTRs or in regions of no significant homology. Xpression Primer will BLAST your sequences, automatically interpret the results and design highly specific primers. The tagged inner primer pair amplifies the PCR product generated by the outer pair with little or no non-coding regions. You can also choose from a list of alternate primers to better meet specific experimental needs. Sequencing for Product Verification Made Easy The versatile algorithm of Xpression Primer can design optimal sequencing primers for multiple sequences in a single run. It picks forward primers across the amplicon at a specified interval and reverse primers staggered on the opposite strand. You can export the results for several popular well plate configurations. Generate Precise Transcripts Design primers to generate sense or antisense transcripts for in vitro expression studies. Web Savvy Xpression Primer searches Entrez and downloads batches of ORF sequences directly into the program. Activate the program following these steps: - Install the Xpression Primer demo from the CD - Launch the program - Click Evaluate on the first window that opens - Request an evaluation key from us - The demo will transform to a fully functional product Learn to use Xpression Primer - Xpression Primer Multimedia Tutorial is included in the CD Order on-line - www.PremierBiosoft.com - E-mail: sales@PremierBiosoft.com - Phone: 650-856-2703, Fax: 650-618-1773 Other bioinformatics tools from Premier Biosoft International AlleleID Array Designer Beacon Designer PREMIER Biosoft International: Visit us at www.PremierBiosoft.com Primer Premier SimGlycan SimVector 3786 Corina Way, Palo Alto, CA 94303-4504 Ph: 650-856-2703, E-mail: support@PremierBiosoft.com, Fax: 650-618-1773 M G B C TG C G C C G A G C C C ATC C ATG C G C C TG C G C C G A G C C ATG C C G TG C C C G C GC C G A G C C C ATTA C C TG C A C C TG C G C TG C G C C C ATC C C C TG C G C TG C G C C G A G C ATTC C ATG C C C TG C G C C G A G C ATG A C C TG C G C C TG G C C G A G C C C ATTA G C TG T C C TG C G C G A G C C C ATC C A TG C G C C TG C G C C G A G C C C ATG C C ATG C G C TG C G C C G A G C C C ATAT G ATG C T C C G C G C C G A G C C C ATC C C C TG C G C C TG C G C G A G C C C ATG A C G TG C A AlleleID 4 A comprehensive tool designed to address the challenges of pathogen detection, bacterial identification, species identification and taxa discrimination using TaqMan®, molecular beacon and microarray assays (for Windows & Mac) A rray Des gner T G G AT A T G C C T A G A G A TG GC T A C A G C T C C T A T A GC G CA A T A A T 4 T G For fast and efficient design of specific oligos for whole genome arrays, tiling arrays and resequencing arrays (for Win & Linux) CG T AA G G AT CG AG C TCG A G AT A TC T AC C AG A G AT AA AG T TC TC TC C TC CT C GC CHAC CG TAT C A G TA C H A TT CG TT A A A LN A T C G T AT A TG CG B De ae cs oi gn n e r 6 Design SYBR® Green primers, TaqMan® probes, MethyLight TaqMan® probes, LNA spiked TaqMan® probes, FRET probes or molecular beacons for robust amplification and fluorescence in real time PCR (for Win & Mac) 5 premier AatI EcoRI BamHI A comprehensive primer design tool (for Win) Ne uAca2-3Galb1-4GlcNAcb1 Ne uAca2-3Galb1 SimGlycan 6 Gal NAca -Thr 3 A comprehensive glycan mass fingerprinting tool (for Win) S mVector 4 T3 Amp T7 pGFP-NI GFP An exceptional tool for drawing publication, vector catalog quality maps and designing cloning experiments (for Win & Mac) Use AlleleID to design probes and primers for pathogen detection, bacterial identification, species identification and taxa discrimination using qPCR and microarray assays. Align sequences to locate differences in DNA and to find conserved regions. Then proceed to design oligos to amplify and detect only the species or strains of interest from the mix. To design taxa specific or cross species assays, design the least number of oligos to identify a group of genes of interest. AlleleID supports assays for selectively amplifying cDNA from gDNA and designs oligos to detect splice variants using microarrays. Design efficient, specific oligos for SNP genotyping and expression microarrays. Start with a list of sequences or the whole genome sequence to study entire organisms effortlessly. Array Designer automatically interprets BLAST results to design specific oligos. Design tiling arrays by avoiding repetitive regions, spot every base of a genomic sequence, to characterize regulatory elements, or to study epigenetic modifications, methylation patterns or protein binding sites. Or design resequencing arrays, far less expensive than the capillary or dideoxy sequencing alternatives, for rapid and accurate populations genotyping studies. Beacon Designer designs oligos for SYBR® Green, TaqMan®, LNA spiked TaqMan®, MethyLight, molecular beacon, NASBA® and FRET assays. Highly specific primers are designed by avoiding cross homologies found by automatically interpreting BLAST search results. Primer efficiency is enhanced by avoiding template secondary structures identified by connecting to the Mfold server. Beacon Designer can be used to evaluate pre-designed assays and designing compatible oligos to extend the designs. It can design beacons and TaqMan® for single tube multiplex experiments. Primer Premier is a program for PCR, hybridization, sequencing, nested and multiplex primer design. It uses ClustalW multiple sequence alignment to design cross species primers automatically in regions of low degeneracy. Primer Premier reverse translates amino acid sequences to design degenerate primers. Designing allele specific primers and primers for site-directed mutagenesis is made easy. Sequences can be analyzed for restriction enzymes and motifs using extensive built-in databases. SimGlycan matches the MS/MS spectra with its own comprehensive, robust and annotated database to predict the structure of the glycan. It generates a list of all probable glycans saving time, money and the frustration of laborious work. Each structure is scored to help judge which results closely match the experimental data. Alongside the probable glycan structure important information such as the glycan class, reaction, pathway and enzyme is also made available. SimVector is a web savvy program equipped with all the drawing tools necessary to publish engineered plasmid catalogs, as well as to simulate and draw plasmid constructs for Gateway®, TA, Restriction or other popular cloning techniques. Export images in vector graphic format to produce the highest quality images for presentations via MS PowerPoint or publication in journals or vector catalogs via Adobe Illustrator. Alternatively, choose popular bitmap formats or export in ready-to-host web pages. With its powerful drawing features, it is easy to enhance vector maps with patterns, colors, fills, curved text and annotations. PREMIER Biosoft International: Visit us at www.PremierBiosoft.com 3786 Corina Way, Palo Alto, CA 94303-4504 Ph: 650-856-2703, E-mail: support@PremierBiosoft.com, Fax: 650-618-1773

 or as ready-to-host web pages.
S mVector 3
T3
or as ready-to-host web pages.
S mVector 3
T3