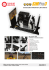Morpheus: a user-friendly modeling environment for multiscale
Transcription
Morpheus: a user-friendly modeling environment for multiscale
Morpheus: a user-friendly modeling environment for multiscale multicellular systems biology Walter de Back Jörn Starruß Lutz Brusch Andreas Deutsch Center for High Performance Computing, Technische Universität Dresden, 01062 Dresden, Germany Contact: walter.deback@tu-dresden.de Website: http://imc.zih.tu-dresden.de/wiki/morpheus Introduction Usability Morpheus is a modeling environment for the simulation and integration of cellbased models with ordinary differential equations and reaction-diffusion systems (Starruß et al., Bioinformatics, 2014). ✦ Modeling without programming Morpheus separates modeling from programming by MorpheusML: a novel domain-specific SBML-like mark-up language for multicellular systems biology. It allows rapid model development of multi-scale models in biological terms and mathematical expressions rather than programming code. Its graphical user interface supports the entire workflow from model construction and simulation to visualization, archiving and batch processing. ✦ Automated model integration It automates model integration linking models of cellular, intra-cellular and extra-cellular dynamics. ✦ Graphical work-flow tools The graphical interface provides tools for rapid model development, simulation, batch processing and archiving. Morpheusʼ graphical user interface Graphical user interface ✦ Model editor MorpheusML is a SBML-like language for multicellular systems biology. The markup language separates modeling from programming and enables users to describe computational models in biological and mathematical terminology rather than programming code. Rapid model development with add/ remove, copy/paste, disable/enable model elements. ✦ Job scheduler Scheduling of multithreaded and parallel simulations, on both local and remote computing resources. Mathematical expressions (functions, differential equations and events) are specified in familiar infix notation. ✦ Simulation archive Browsable archive of simulation models with results, allows old models to be restored. ✦ High performance computing Simulation, visualization and analysis Support for high performance computing with batch systems like LSF and SLURM. Syncing results back to local computer via sftp. ✦ Batch processing Parameter exploration by creating sequences of simulations with different parameter sets. MorpheusML Examples, documentation and downloads for Linux, Mac OSX and Windows at: http://imc.zih.tu-dresden.de/wiki/morpheus MorpheusML enables model integration by automatically resolving dependencies between symbolic identifiers to determine e.g. the order in which model components must be updated. Key references J. Starruß, W. de Back, L. Brusch and A. Deutsch (2014). Morpheus: a user-friendly modeling environment for multiscale and multicellular systems biology. Bioinformatics, btt772. ✦ A. Köhn-Luque, W. de Back, et al. (2013) Dynamics of VEGF MatrixRetention in Vascular Network Patterning. Physical Biology, 10:066007. ✦ W. de Back, R. Zimm, L. Brusch (2013) Transdifferentiation of Pancreatic Cells by Loss of Contact-mediated Signaling. BMC Systems Biology, 7:77. ✦ W. de Back, J. X. Zhou, L. Brusch (2012) On the Role of Lateral Stabilization during Early Patterning in the Pancreas. J. R. Soc. Interface 10(79):20120766. ✦ Model description in MorpheusML Software Modeling and simulation ✦ Differential equations Systems of ordinary, stochastic and delay differential equations can represent temporal dynamics such as gene regulation, metabolism or signaling and can be imported from SBML models available in repositories such as the BioModels database. ✦ Reaction-diffusion ✦ Cell-based modeling Cells can be represented as point-like objects or with explicit 2D or 3D cell shapes, e.g. imported from TIFF microscopy images. Interactions are specified between individual discrete cells. Cell motility and biophysical constraints may be added using the cellular Potts Examples on website and GUI model (CPM) framework. Reaction-diffusion systems can be used to model e.g. the spatial distribution of morphogens and binding/unbinding to extracellular matrix components. The spatial domain may be imported from multistack images. ✦ Multiscale multicellular models To study feedbacks between various levels of biological organization, models can be constructed that couple cellular dynamics with intra- and/or extracellular dynamics. Morpheus is a self-contained application for Linux, Mac OSX and Windows that consists of two stand-alone programs: a C++-based simulator and a Qt-based graphical user interface. It uses muparser to evaluate math expressions, eigen for matrix operations, openMP for parallel computing, libTIFF for import/export of 3D images, SQLite for archiving, and gnuplot as data visualization back-end. Morpheus features a plug-in architecture that allows extensions to be written in C++. This transforms the current binary application into a flexible framework following the planned open-source release. The development of this software is supported by the German Ministry for Education and Research (BMBF) as part of the Virtual Liver Network.